Structure of IDP91417
1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
Edit deposit information
- CSGID target
- IDP91417
- PDB Id
- 3V4G (NCBI MMDB)
- Authors
- 'A.S.Halavaty,G.Minasov,E.Filippova,L.Shuvalova,J.Winsor,I.Dubrovska,S.Peterson,W.F.Anderson,Center For Structural Genomics Of Infectious Diseases (Csgid)'
- Responsible person
- Andrei Halavaty
- Responsible lab
- Northwestern University
- Deposition Date
- Dec 14, 2011
- Release Date
- Jan 04, 2012
Annotation
- Description
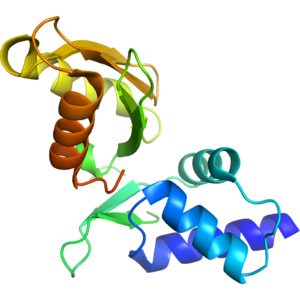
- The 1.6 Å resolution structure of the Vibrio vulnificus arginine repressor was determined by molecular replacement using two models: (i) C-terminal domain of Escherichia coli arginine repressor/L-arginine complex (PDB code 1XXB); and (ii) the Reovirus outer capsid protein Sigma (3PDB code 1FN9). The first model was successful in finding a structure solution for a C-terminal domain of the protein, whereas the second structure was useful in finding solution for a N-terminal domain of the arginine repressor. The asymmetric unit is comprised of a single polypeptide chain with an alpha/beta fold. The N-terminal domain has three antiparallel helices and a two-stranded antiparallel β-sheet. The C-terminal domain contains a four-stranded antiparallel β-sheet and three antiparallel helices. Residues 71-76 that link the two domains are disordered in the structure. The C-terminal domain sits on a 3-fold crystallographic axis creating a trimer (buried surface area is 4380 Å2) with another two symmetry-related molecules. Another possible oligomer in the crystal is a hexamer (buried surface area is 14430 Å2) that is formed from two trimers positioned back-to-back along the aforementioned axis.
- Functional assignment
- DNA BINDING PROTEIN
Ligands
| Ligand code | Name | Ligand type |
|---|
Structure information
Unit cell parameters
- Space Group
- P 63 2 2
- Unit Cell
-
a=73.31Å, b=73.31Å, c=118.60Å
α=90.00, β=90.00, γ=120.00 - Solvent content
- Matthews coefficient
- Resolution range
- 29.65-1.60Å (1.64-1.60Å)
- Rall(%)
- 18.2
- Rwork(%)
- 18.2 (21.8)
- Rfree(%)
- 19.8 (27.0)
- Num. observed reflections
- 25557 (1828)
- Num. Rfree reflections
- 1303 (87)
- Completeness(%)
- 100.0 (100.0)
- Num Atoms
- 1257
- Num Waters
- 176
- Num Hetatoms
- 184
- Model mean isotropic B factor
- 23.940Å2
- RMSD bond length
- 0.008Å
- RMSD bond angle
- 1.602°
- Filename uploaded
- 3V4G.pdb (uploaded on Jan 10, 2012 12:27 PM)
- Inserted
- Dec 26, 2011