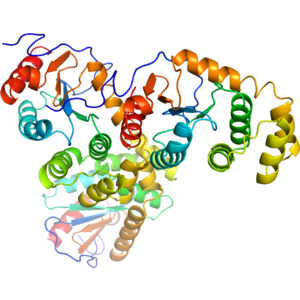
Structure of IDP91802
Crystal structure of Disulfide Bond Oxidoreductase DsbA1 from Legionella pneumophila
Edit deposit information
- CSGID target
- IDP91802
- PDB Id
- 4JRR (NCBI MMDB)
- Authors
- I.A.Shumilin,M.Jameson-Lee,M.Cymborowski,M.J.Domagalski,O.Chertihin,Z.Z.Kpadeh,A.J.Yeh,P.S.Hoffman,W.F.Anderson,W.Minor,Center For Structural Genomics Of Infectious Diseases (Csgid)
- Responsible person
- Igor Shumilin
- Responsible lab
- University of Virginia
- Deposition Date
- Mar 21, 2013
- Release Date
- Apr 24, 2013
Annotation
- Description
- The DSB oxidoreductase family is part of the thioredoxin (TRX) super family of proteins, which are defined by the presence of one or more thioredoxin folds. These enzymes participate in disulfide bond formation through a conserved Cys-X-X-Cys (CXXC) active site motif, and can either reduce or oxidize target substrates. DsbA catalyzes the formation of consecutive disulfide bonds in nascent polypeptides entering the periplasm.
- Functional assignment
- Disulfide Bond Oxidoreductase
Ligands
| Ligand code | Name | Ligand type |
|---|---|---|
| GOL | glycerol | crystallization |
| 175 | 3,5-dihydro-5-methylidene-4h-imidazol-4-on |
Structure information
Unit cell parameters
- Space Group
- P 1 21 1
- Unit Cell
-
a=55.88Å, b=80.14Å, c=66.26Å
α=90.00, β=92.86, γ=90.00 - Solvent content
- Matthews coefficient
- Resolution range
- 36.98-1.88Å (1.93-1.88Å)
- Rall(%)
- 16.4
- Rwork(%)
- 16.2 (24.1)
- Rfree(%)
- 20.5 (30.7)
- Num. observed reflections
- 47488 (3486)
- Num. Rfree reflections
- 2421 (154)
- Completeness(%)
- 99.9 (98.8)
- Num Atoms
- 4544
- Num Waters
- 376
- Num Hetatoms
- 414
- Model mean isotropic B factor
- 32.920Å2
- RMSD bond length
- 0.019Å
- RMSD bond angle
- 1.808°
- Filename uploaded
- hkl_refine_53.pdb (uploaded on Mar 21, 2013 7:42 PM)
- Inserted
- Mar 21, 2013